Comparison of Three Damping Cases
The project file for this example may be viewed/run in FLAC3D.[1] The main data file used is shown at the end of this example.
The i Integration of Damping Schemes and Nonlinear Material Models for Geo-materials section in i Dynamic Damping presents three damping schemes:
The energy dissipation for these three damping cases is compared by exercising the equations for \(G/G_{max}\) and \(D\) over a cyclic strain range. A FISH function, damp, listed below, performs this exercise over a cyclic shear strain, \(\gamma_c\), from 0.0001 to 4.0. The value for \(\gamma_m\) is set to 0.01 and the value for \(\gamma_{ref}\) is set to 0.02. The results for \(G/G_{max}\) versus log \(\gamma_c/\gamma_m\), based upon Equations (71), (77), (90), and (91), are plotted in Figures 1. The results for \(D\) versus log \(\gamma_c/\gamma_m\), based upon Equations (76), (85), and (97), are plotted in Figure 2.
The inclusion of hysteretic damping is shown to reduce the shear modulus from the initial value of \(G_{max}\), and increase the damping ratio (compared to the elastic-only response). The damping ratio increases monotonically with shear strain amplitude and approaches the asymptotic value of \(2 / \pi\) for all three cases.
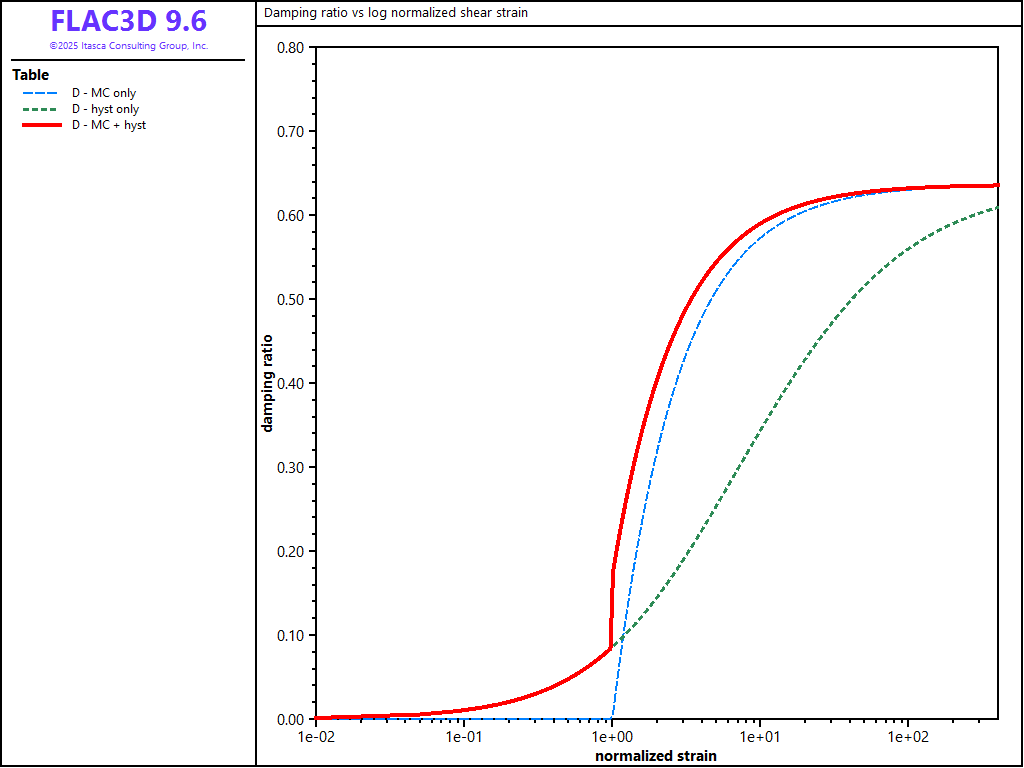
Figure 1: Normalized shear modulus vs. log normalized shear strain for three damping cases in FLAC3D.
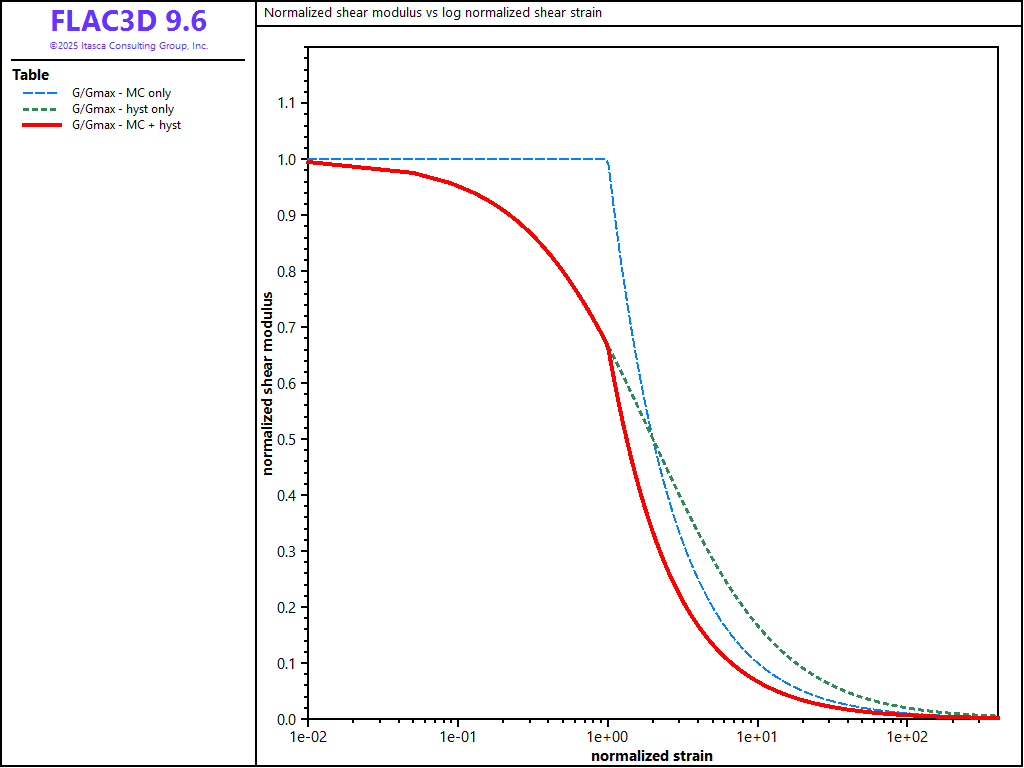
Figure 2: Damping ratio vs. log normalized shear strain for three damping cases in FLAC3D.

Figure 3: Normalized shear modulus vs. log normalized shear strain for three damping cases in FLAC2D.

Figure 4: Damping ratio vs. log normalized shear strain for three damping cases in FLAC2D.
Data File (FLAC3D)
Damping-Compare.dat
;---------------------------------------------------------------------
; Compare damping
; Driver file
;---------------------------------------------------------------------
model new
fish automatic-create off
; assume Gmax = 1.
fish define damp
; strain range
local r1 = 0.0001 ; min
local r2 = 4. ; max
local rm = 0.01
local rref = 0.02
local np = 10000
local rinc = (r2-r1)/float(np)
local r = r1
loop local n (1,np)
local rrat = r / rm
if r <= 0. then
local ghyst = 1.
local dhyst = 0.
else
local tau = r/(1.+r/rref)
ghyst = tau/r
local rf = r/rref
local rf2 = rf*rf
local rln = rf - math.ln(1.+rf)
dhyst = 2./math.pi*(2.*(1.+rf)/rf^2*rln-1.)
endif
if r < rm then
local gmohr = 1.
local dmohr = 0.
else
gmohr = rm/r
dmohr = 2.*(r-rm)/(math.pi*r)
endif
if r <= rm then
local gcomb = 1./(1.+r/rref)
else
gcomb = 1./((1.+rm/rref)*r/rm)
endif
if r <= rm then
local dcomb = dhyst
else
rf = rm/rref
rf2 = rf*rf
rln = rf - math.ln(1.+rf)
dcomb = 2./math.pi*(2.*(1.+rf)/rf^2*rln-1.)*rref/r
dcomb = dcomb + 2./math.pi*(r-rm)/r
endif
rrat = rrat
table(11,rrat) = gmohr
table(21,rrat) = dmohr
table(12,rrat) = ghyst
table(22,rrat) = dhyst
table(13,rrat) = gcomb
table(23,rrat) = dcomb
r = r + rinc
endloop
end
[damp]
Data File (FLAC2D)
Damping-Compare.dat
;---------------------------------------------------------------------
; Compare damping
; Driver file
;---------------------------------------------------------------------
model new
fish automatic-create off
;
; assume Gmax = 1.
;
fish define damp
; strain range
local r1 = 0.0001 ; min
local r2 = 4. ; max
local rm = 0.01
local rref = 0.02
local np = 10000
local rinc = (r2-r1)/float(np)
local r = r1
loop local n (1,np)
local rrat = r / rm
if r <= 0. then
local ghyst = 1.
local dhyst = 0.
else
local tau = r/(1.+r/rref)
ghyst = tau/r
local rf = r/rref
local rf2 = rf*rf
local rln =rf - math.ln(1.+rf)
dhyst = 2./math.pi*(2.*(1.+rf)/rf^2*rln-1.)
endif
if r < rm then
local gmohr = 1.
local dmohr = 0.
else
gmohr = rm/r
dmohr = 2.*(r-rm)/(math.pi*r)
endif
if r <= rm then
local gcomb = 1./(1.+r/rref)
else
gcomb = 1./((1.+rm/rref)*r/rm)
endif
if r <= rm then
local dcomb = dhyst
else
rf = rm/rref
rf2 = rf*rf
rln = rf - math.ln(1.+rf)
dcomb = 2./math.pi*(2.*(1.+rf)/rf^2*rln-1.)*rref/r
dcomb = dcomb + 2./math.pi*(r-rm)/r
endif
rrat = math.log(rrat)
table(11,rrat) = gmohr
table(21,rrat) = dmohr
table(12,rrat) = ghyst
table(22,rrat) = dhyst
table(13,rrat) = gcomb
table(23,rrat) = dcomb
r = r + rinc
endloop
end
[damp]
program return
Endnote
⇐ Velocity Wave with Sharp Front, Artificial Viscosity (FLAC2D) | Spatial Variation in Damping (FLAC3D) ⇒
| Was this helpful? ... | Itasca Software © 2026, Itasca | Updated: Feb 07, 2026 |
